General Introduction
Artificial intelligence (AI) is increasingly addressing major limitations in biomedical research, from empirical nanoparticle screening to the complex interpretation of protein function, omics datasets, and cellular behaviors. Traditional experimental workflows often struggle with the scale, cost, and biological variability inherent to modern therapeutic development. Recent AI advances now offer predictive, mechanistic, and integrative models capable of operating across multiple biological levels, from molecular interactions to cell systems and virtual patients 1, 2, 3.
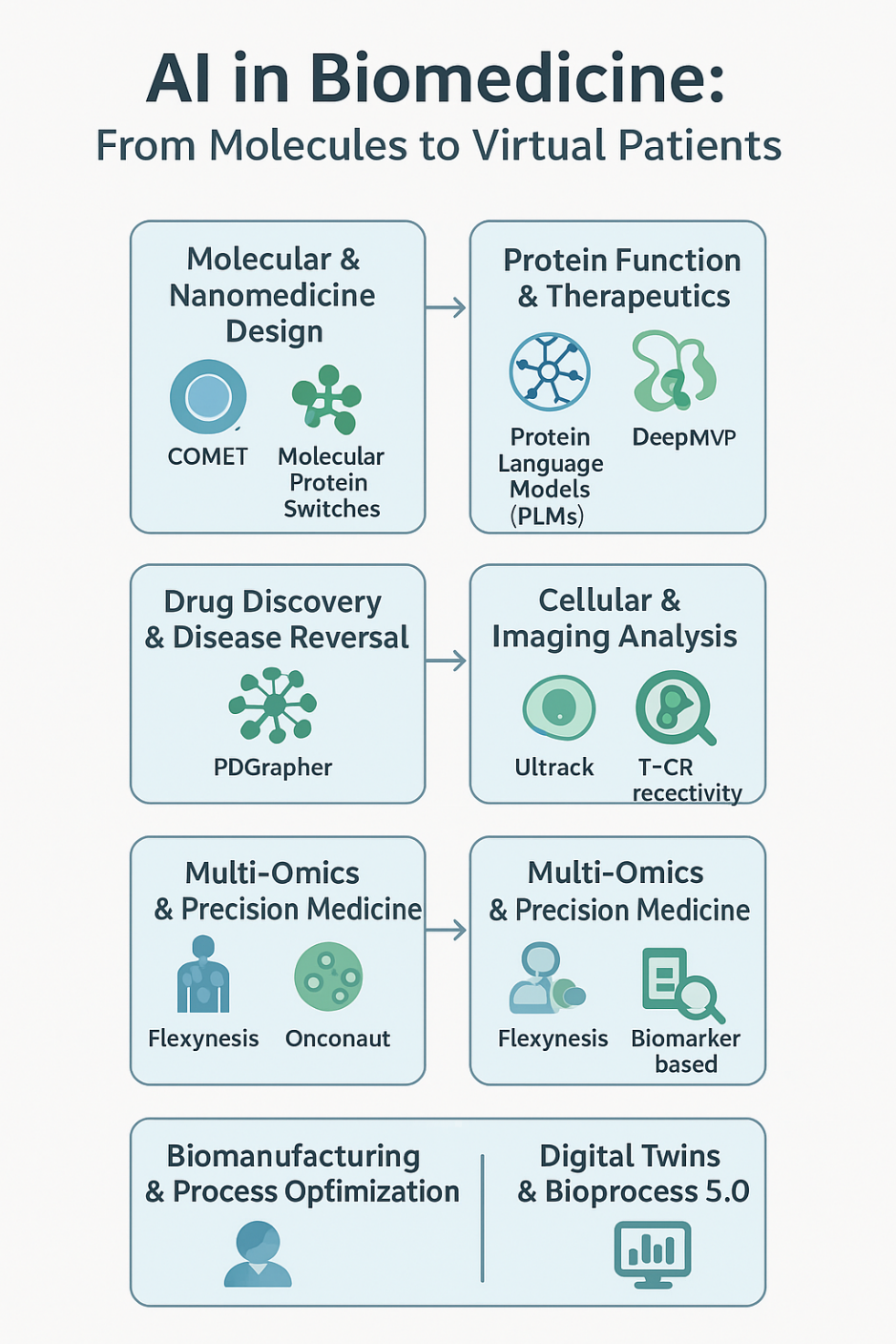
These innovations include tools that accelerate:

1. Molecular and Nanomedicine Design
AI is enabling smarter design of nanomedicines and molecular interventions
• COMET (MIT) – At the molecular level, AI is enabling smarter nanomedicine design. Researchers at MIT have developed COMET, an AI model that predicts optimal lipid nanoparticle (LNP) formulations for mRNA delivery. Trained on over 3,000 experimentally tested LNPs, COMET can anticipate performance, optimize delivery to specific cell types, and incorporate new polymer–lipid hybrid materials. By shifting from slow trial-and-error screening to AI-guided design, COMET is accelerating the development of RNA therapies for metabolic diseases 4.
• Baker Lab Molecular Protein Switches (UW)– At the protein interaction level, the lab of David Baker at the University of Washington has designed molecular on/off switches that control protein activity with unprecedented speed. These switches accelerate protein dissociation up to 6,000-fold, enabling precise temporal control. Applied to interleukin-2 (IL-2) for cancer immunotherapy, these switches can temporarily silence immune activation, improving safety. They also enhance biosensor responsiveness, exemplified by a SARS-CoV-2 sensor responding 70 times faster than previous protein-based tests 5.
2. Protein Function and Therapeutics
AI is advancing our understanding and engineering of proteins.
• Protein Language Models (PLMs, MIT) – In parallel, AI is unveiling the inner workings of protein language models (PLMs), which analyze proteins like language models process text. MIT researchers led by Bonnie Berger applied sparse autoencoders to “disentangle” neural networks, linking individual neurons to biological features. This interpretability is crucial for applications such as antibody engineering and drug–target prediction, allowing scientists to understand and trust AI-driven insights 6.
• DeepMVP (Baylor College of Medicine) – Complementing this, researchers at Baylor College of Medicine developed DeepMVP, an AI tool that predicts post-translational modification (PTM) sites on proteins and how mutations disrupt them. PTMs are crucial for protein function, and their disruption can drive disease. DeepMVP, trained on nearly 400,000 curated PTM sites, achieves high accuracy in predicting PTM location and direction of change, aiding research into cancer, neurological, and cardiovascular diseases while informing potential drug targets 7.
• BATMAN (Cold Spring Harbor Lab) – AI is further improving immunotherapy safety. At Cold Spring Harbor Laboratory, researchers developed BATMAN, a model predicting T cell receptor (TCR) cross-reactivity. Trained on over 20,000 TCR–peptide interactions (BATCAVE dataset), BATMAN identifies how peptide mutations affect TCR activation, helping to minimize off-target immune responses in engineered T cell therapies 8.
3. Drug Discovery and Disease Reversal
AI reshapes drug discovery by identifying interventions that reverse disease states.
• PDGrapher (Harvard Medical School) – At Harvard Medical School, the PDGrapher model predicts gene targets or drug combinations that can reverse disease states in cells. By using graph neural networks to map interactions between genes, proteins, and pathways, PDGrapher identifies interventions that are likely to restore healthy cellular function. Tested across multiple cancer datasets, the model successfully predicted known therapeutic targets and revealed novel candidates, offering faster, more precise paths for complex disease treatment and personalized therapies 9.
4. Cellular and Imaging Analysis
Cutting-edge AI technologies are transforming the way researchers visualize and analyze cellular processes at high resolution
• Ultrack & inTRACKtive (Chan Zuckerberg Biohub) – For understanding cellular behavior, the Chan Zuckerberg Biohub introduced Ultrack and inTRACKtive, tools that combine deep learning and interactive 3D visualization to track cell division, migration, and organization in massive time-lapse datasets. These tools allow scientists to explore complex developmental and disease-related processes with unprecedented clarity and scale 10.
5. Multi-Omics and Precision Medicine
AI integrates multi-dimensional patient data to guide personalized treatment.
• Flexynesis (Max Delbrück Center) – precision oncology benefits from AI through tools like Flexynesis, a deep-learning system that integrates multi-omics, clinical data, and imaging to guide personalized cancer treatment. By predicting tumor subtypes, drug responses, and survival probabilities, Flexynesis empowers clinicians to select tailored therapies, complementing existing biomarker-based approaches 11.
• Onconaut (previous tool, complementary to Flexynesis) – Biomarker-based AI for clinical decision support in oncology 12.
6. Biomanufacturing and Process Optimization
AI optimizes production and scaling of complex therapies.
• Digital Twins & Bioprocess 5.0 (ELEM Biotech, Cytiva, Merck) – At the systems level, digital twins and Bioprocess 5.0 are transforming biopharma and personalized medicine. Companies like ELEM Biotech and Cytiva are creating virtual patients and AI-driven manufacturing simulations to optimize clinical trials and scale production of complex therapies such as CAR T cells and antibody-drug conjugates. These approaches integrate AI, automation, and human-centered decision-making to ensure reproducibility, safety, and sustainability 13.
Conclusion
The growing ecosystem of AI models represents a significant shift in how modern therapeutics are discovered, optimized, and translated. By replacing slow trial-and-error processes with predictive and interpretable computational frameworks, AI accelerates the design of nanomedicines, proteins, and targeted therapies while improving safety and mechanistic understanding. The observed progress across domains, ranging from improved mRNA delivery systems to safer immunotherapies, more robust disease reversal predictions, and refined oncology decision support, highlights the broad impact of AI on biomedical innovation.
As AI continues to integrate multi-omics data, imaging, and virtual patient simulations, it is poised to become a central driver of personalized medicine and bioprocess automation. Future developments will focus on reinforcing interpretability, ensuring model robustness, and enabling seamless clinical integration. Overall, AI-guided approaches are transforming the therapeutic pipeline and hold strong promise for delivering more effective and patient-tailored healthcare solutions.
References:
(1) Rashid Bhat, A.; Ahmed, S. Artificial Intelligence ( AI ) in Drug Design and Discovery : A Comprehensive Review. Silico Res. Biomed. J. 2025, 1 (August), 1–13. https://doi.org/https://doi.org/10.1016/j.insi.2025.100049.
(2) Liu, J.; Cen, X.; Yi, C.; Wang, F.; Ding, J.; Cheng, J.; Wu, Q.; Gai, B.; Zhou, Y.; He, R.; Gao, F.; Li, Y. Challenges in AI-Driven Biomedical Multimodal Data Fusion and Analysis. Genomics. Proteomics Bioinformatics 2025, 23 (1), 1–20. https://doi.org/https://doi.org/10.1093/gpbjnl/qzaf011.
(3) Han, S.; Wu, J. Artificial Intelligence (AI) Meets Biomaterials and Biomedicine. Smart Mater. Med. 2024, 5, 251–255. https://doi.org/https://doi.org/10.1016/j.smaim.2024.03.001
(4) Chan A., Kirtane A. R., Qu Q. R., Huang X., Woo J., Subramanian D. A., Dey R., Semalty R., Bernstock J. D., Ahmed T., Honeywell R., Hanhurst Ch., Becdach I. D., Prizant L .S., Brown A. K., Song H., Cobb J. L., DeRidder L. B., Santos B., Jimenez M., Sun M., T. G. Designing Lipid Nanoparticles Using a Transformer-Based Neural Network. Nat. Nanotechnol. 2025, 20, 1491–1501. https://doi.org/https://doi.org/10.1038/s41565-025-01975-4.
(5) Broerman A. J., Pollmann Ch., Zhao Y., Lichtenstein M. A., Jackson M. D., Tessmer M. H., Ryu W. H., Ogishi M., Abedi M. H., Sahtoe D. D., Allen A., Kang A., De La Cruz J., Brackenbrough E., Sankaran B., Bera A. K., Zuckerman D. M., Stoll S., Garcia K. Ch., B. D. Design of Facilitated Dissociation Enables Timing of Cytokine Signalling. Nature 2025, 647, 529–535. https://doi.org/https://doi.org/10.1038/s41586-025-09549-z
(6) Gujral, O.; Bafna, M.; Alm, E.; Berger, B. Sparse Autoencoders Uncover Biologically Interpretable Features in Protein Language Model Representations. Appl. Math. 2025, 122 (34), 1–12. https://doi.org/10.1073/pnas.2506316122/-/DCSupplemental.Published.
(7) Wen, B.; Wang, C.; Li, K.; Han, P.; Holt, M. V; Savage, S. R.; Lei, J. T.; Dou, Y.; Shi, Z.; Li, Y.; Zhang, B. DeepMVP:Deep Learning Models Trained on High-Quality Data Accurately Predict PTM Sites and Variant-Induced Alterations. Nat. Methods 2025, 22 (September), 1857–1867. https://doi.org/https://doi.org/10.1038/s41592-025-02797-x P.
(8) Banerjee, A.; Pattinson, D. J.; Wincek, C. L.; P., B.; Chapin, S. R.; Navlakha, S.; Meyer, H. V. BATMAN:Improved T Cell Receptor Cross-Reactivity Prediction Benchmarked on a Comprehensive Mutational Scan Database. bioRxiv (The Prepr. Serv. Biol. 2024, 1–18. https://doi.org/https://doi.org/10.1101/2024.01.22.576714.
(9) Gonzalez, G.; Lin, X.; Herath, I.; Veselkov, K.; Bronstein, M.; Zitnik, M. Combinatorial Prediction of Therapeutic Perturbations Using Causally Inspired Neural Networks. Nat. Biomed. Eng. Artic. 2025, 1–30. https://doi.org/https://doi.org/10.1038/s41551-025-01481-x
(10) Bragantini, J.; Theodoro, I.; Zhao, X.; Huijben, T. A. P. M.; Hirata-Miyasaki, E.; Vijaykumar, S.; Balasubramanian, A.; Lao, T.; Agrawal, R.; Xiao, S.; Lammerding, J.; Mehta, S.; Falcão, A. X.; Jacobo, A.; Lange, M.; Royer, L. A. Ultrack:Pushing the Limits of Cell Tracking across Biological Scales. Nat. Methods 2025, 22, 2423–2436. https://doi.org/https://doi.org/10.1038/s41592-025-02778-0.
(11) Uyar, B.; Savchyn, T.; Nilchi, A. N.; Sarigun, A.; Wurmus, R.; Shaik, M. M.; Gruning, B.; Franke, V.; Akalin, A. Flexynesis: A Deep Learning Toolkit for Bulk Multi-Omics Data Integration for Precision Oncology and Beyond. Nat. Commun. 2025, 16 (8261), 1–18. https://doi.org/https://doi.org/10.1038/s41467-025-63688-5
(12) Deep Learning Integrates Multi-Omics for Precision Oncology Decision Making. https://www.genengnews.com/topics/artificial-intelligence/deep-learning-integrates-multi-omics-for-precision-oncology-decision-making/.
(13) Fay Lin, P. Bioprocess 5.0: Straddles the Human-AI Divide to Scale Next-Gen Therapies. https://www.genengnews.com/topics/bioprocessing/bioprocess-5-0-straddles-the-human-ai-divide-to-scale-next-gen-therapies/ (accessed 2025-12-08).